Community Highlights
Scalable Molecular Dynamics with Work Queue at UT-Austin
The Biomolecular Engineering Lab at UT-Austin routinely requires large scale molecular dynamics for predicting ligand-protein binding affinity. The lab makes use of the Cooperative Computing Tools to build and run a variety of distributed applications on their 124 node, 75 GPU cluster. Custom Work Queue applications are run on the cluster for months at a time to generate large amounts of ab-initio data to parameterize the AMOEBA model for small molecules, and perform single-point computations via Poltype 2. In addition, the lab makes use of the ForceBalance application built on Work Queue for liquid property fitting for Van der Waals parameter refinement.
OpenTopography + EEMT + Makeflow
The OpenTopography service provides online access to geospatial data and computational tools in support of earth sciences. The Effective Energy and Mass Transfer (EEMT) tool allows for computations of energy transfer in the Earth's critical zone, taking into account topography, vegetation, weather, and so forth. To scale these computations up to large clusters, the CCL's Makeflow and Work Queue frameworks are employed to construct large scale parallel workflows at the touch of a button from the OpenTopography website.Source: Tyson Swetnam, University of Arizona
Analyzing Agriculture with Work Queue
The Field Scanalyzer at the University of Arizona is a massive robot that uses sensors, cameras, and GPS devices to collect vast quantities of agricultural data from crop fields. In the background, distributed computing and deep learning techniques are used to understand and improve agricultural efficiencies in hot, dry, climates. Processing all this data requires reliable computation on large clusters: the PhytoOracle software from the Lyons Lab at UA makes this possible, building on the Work Queue software from the Cooperative Computing Lab at Notre Dame.
- Source: Eric Lyons University of Arizona
IceCube Flies with Parrot and CVMFS
IceCube is a neutrino detector built at the South Pole by instrumenting about a cubic kilometer of ice with 5160 light sensors. The IceCube data is analyzed by a collaboration of about 300 scientists from 12 countries. Data analysis relies on the precise knowledge of detector characteristics, which are evaluated by vast amounts of Monte Carlo simulation. On any given day, 1000-5000 jobs are continuously running.
Recently, the experiment began using Parrot to get their code running on GPU clusters at XSEDE sites (Comet, Bridges, and xStream) and the Open Science Grid. IceCube relies on software distribution via CVMFS, but not all execution sites provide the necessary FUSE modules. By using Parrot, jobs can attach to remote software repositories without requiring special privileges or kernel modules.
- Courtesy of Gonzalo Merino, University of Wisconsin - Madison
CCL Workshop 2016
 The 2016 CCL Workshop on Scalable Scientific Computing was held on October 19-20 at the University of Notre Dame. We offered tutorials on Makeflow, Work Queue, and Parrot. and gave highlights of the many new capabilities relating to reproducibility and container technologies. Our user community gave presentations describing how these technologies are used to accelerate discovery in genomics, high energy physics, molecular dynamics, and more.
The 2016 CCL Workshop on Scalable Scientific Computing was held on October 19-20 at the University of Notre Dame. We offered tutorials on Makeflow, Work Queue, and Parrot. and gave highlights of the many new capabilities relating to reproducibility and container technologies. Our user community gave presentations describing how these technologies are used to accelerate discovery in genomics, high energy physics, molecular dynamics, and more. Everyone got together to share a meal, solve problems, and generate new ideas. Thanks to everyone who participated, and see you next year!
Summer REU Projects in Data Intensive Scientific Computing
We recently wrapped up the first edition of the summer REU in Data Intensive Scientific Computing at the University of Notre Dame. Ten undergraduate students came to ND from around the country and worked on projects encompassing physics, astronomy, bioinformatics, network sciences, molecular dynamics, and data visualization with faculty at Notre Dame.To learn more, see these videos and posters produced by the students:
Simulation of HP24stab with AWE and Work Queue

Work Queue from Raspberry Pi to Azure at SPU
"At Seattle Pacific University we have used Work Queue in the CSC/CPE 4760 Advanced Computer Architecture course in Spring 2014 and Spring 2016. Work Queue serves as our primary example of a distributed system in our “Distributed and Cloud Computing” unit for the course. Work Queue was chosen because it is easy to deploy, and undergraduate students can quickly get started working on projects that harness the power of distributed resources."The main project in this unit had the students obtain benchmark results for three systems: a high performance workstation; a cluster of 12 Raspberry Pi 2 boards, and a cluster of A1 instances in Microsoft Azure. The task for each benchmark used Dr. Peter Bui’s Work Queue MapReduce framework; the students tested both a Word Count and Inverted Index on the Linux kernel source. In testing the three systems the students were exposed to the principles of distributed computing and the MapReduce model as they investigated tradeoffs in price, performance, and overhead.
- Prof. Aaron Dingler, Seattle Pacific University.
Lifemapper analyzes biodiversity using Makeflow and Work Queue
Lifemapper is a high-throughput, webservice-based, single- and multi-species modeling and analysis system designed at the Biodiversity Institute and Natural History Museum, University of Kansas. Lifemapper was created to compute and web publish, species distribution models using available online species occurrence data. Using the Lifemapper platform, known species localities georeferenced from museum specimens are combined with climate models to predict a species’ “niche” or potential habitat availability, under current day and future climate change scenarios. By assembling large numbers of known or predicted species distributions, along with phylogenetic and biogeographic data, Lifemapper can analyze biodiversity, species communities, and evolutionary influences at the landscape level.Lifemapper has had difficulty scaling recently as our projects and analyses are growing exponentially. For a large proof-of-concept project we deployed on the XSEDE resource Stampede at TACC, we integrated Makeflow and Work Queue into the job workflow. Makeflow simplified job dependency management and reduced job-scheduling overhead, while Work Queue scaled our computation capacity from hundreds of simultaneous CPU cores to thousands. This allowed us to perform a sweep of computations with various parameters and high-resolution inputs producing a plethora of outputs to be analyzed and compared. The experiment worked so well that we are now integrating Makeflow and Work Queue into our core infrastructure. Lifemapper benefits not only from the increased speed and efficiency of computations, but the reduced complexity of the data management code, allowing developers to focus on new analyses and leaving the logistics of job dependencies and resource allocation to these tools.
Information from C.J. Grady, Biodiversity Institute and Natural History Museum, University of Kansas.
CMS Analysis on 10K Cores with Lobster
 The CMS physics group at Notre Dame has created Lobster, a data analysis system that runs on O(10K) cores to process data produced by the CMS experiment at the LHC. Lobster uses Work Queue to dispatch tasks to thousands of machines, Parrot with CVMFS to deliver the complex software stack from CERN, XRootD to deliver the LHC data, and Chirp and Hadoop to manage the output data.
By using these technologies, Lobster is able to harness arbitrary machines and bring along the CMS computing environment wherever it goes. At peak, Lobster at ND delivers capacity equal to that of a dedicated CMS Tier-2 facility! (read more here)
The CMS physics group at Notre Dame has created Lobster, a data analysis system that runs on O(10K) cores to process data produced by the CMS experiment at the LHC. Lobster uses Work Queue to dispatch tasks to thousands of machines, Parrot with CVMFS to deliver the complex software stack from CERN, XRootD to deliver the LHC data, and Chirp and Hadoop to manage the output data.
By using these technologies, Lobster is able to harness arbitrary machines and bring along the CMS computing environment wherever it goes. At peak, Lobster at ND delivers capacity equal to that of a dedicated CMS Tier-2 facility! (read more here)
Creating Better Force Fields on Distributed GPUs with Work Queue
ForceBalance is an open source software tool for creating accurate force fields for molecular mechanics simulation using flexible combinations of reference data from experimental measurements and theoretical calculations. These force fields are used to simulate the dynamics and physical properties of molecules in chemistry and biochemistry.The Work Queue framework gives ForceBalance the ability to distribute computationally intensive components of a force field optimization calculation in a highly flexible way. For example, each optimization cycle launched by ForceBalance may require running 50 molecular dynamics simulations, each of which may take 10-20 hours on a high end NVIDIA GPU. While GPU computing resources are available, it is rare to find 50 available GPU nodes on any single supercomputer or HPC cluster. With Work Queue, it is possible to distribute the simulations across several HPC clusters, including the Certainty HPC cluster at Stanford, the Keeneland GPU cluster managed by Georgia Tech and Oak Ridge National Laboratories, and the Stampede supercomputer managed by the University of Texas. This makes it possible to run many simulations in parallel and complete the high level optimization in weeks instead of years.
- Lee-Ping Wang, Stanford University
Work Queue Powers Nanoreactor Simulations
 Lee-Ping Wang at Stanford University, recently published a paper in Nature Chemistry describing his work in fundamental molecular dynamics.
Lee-Ping Wang at Stanford University, recently published a paper in Nature Chemistry describing his work in fundamental molecular dynamics.The paper demonstrates the "nanoreactor" technique in which simple molecules are simulated over a long time scale to observe their reaction paths into more complex molecules. For example, the picture below shows 39 Acetylene molecules merging into a variety of hydrocarbons over the course of 500ps simulated time. This technique can be used to computationally predict reaction networks in historical or inaccessible environments, such as the early Earth or the upper atmosphere.
To compute the final reaction network for this figure, the team used the Work Queue framework to harness over 300K node-hours of CPU time on the Blue Waters supercomputer at NCSA.
Accelerating Protein Folding with Adaptive Weighted Ensemble and Work Queue
 Computational protein folding has historically relied on long-running
simulations of single molecules. Although many such simulations can run
be at once, they are statistically likely to sample the same common
configurations of the molecule, rather than exploring the many possible
states it may have. To address this, a team of researchers from the
University of Notre Dame and Stanford University designed a system that
combined the Adaptive Weighted Ensemble technique to run thousands of short Gromacs and Protomol simulations in parallel with periodic resampling to explore the rich state space of a molecule. Using the Work Queue
framework, these simulations were distributed across thousands of CPUs
and GPUs drawn from the Notre Dame, Stanford, and commercial cloud
providers. The resulting system effectively simulates the behavior of a
protein at 500 ns/hour, covering a wide range of behavior in days
rather than years.
Computational protein folding has historically relied on long-running
simulations of single molecules. Although many such simulations can run
be at once, they are statistically likely to sample the same common
configurations of the molecule, rather than exploring the many possible
states it may have. To address this, a team of researchers from the
University of Notre Dame and Stanford University designed a system that
combined the Adaptive Weighted Ensemble technique to run thousands of short Gromacs and Protomol simulations in parallel with periodic resampling to explore the rich state space of a molecule. Using the Work Queue
framework, these simulations were distributed across thousands of CPUs
and GPUs drawn from the Notre Dame, Stanford, and commercial cloud
providers. The resulting system effectively simulates the behavior of a
protein at 500 ns/hour, covering a wide range of behavior in days
rather than years.- Jesus Izaguirre, University of Notre Dame and Eric Darve, Stanford University
Teaching Distributed Computing with Work Queue
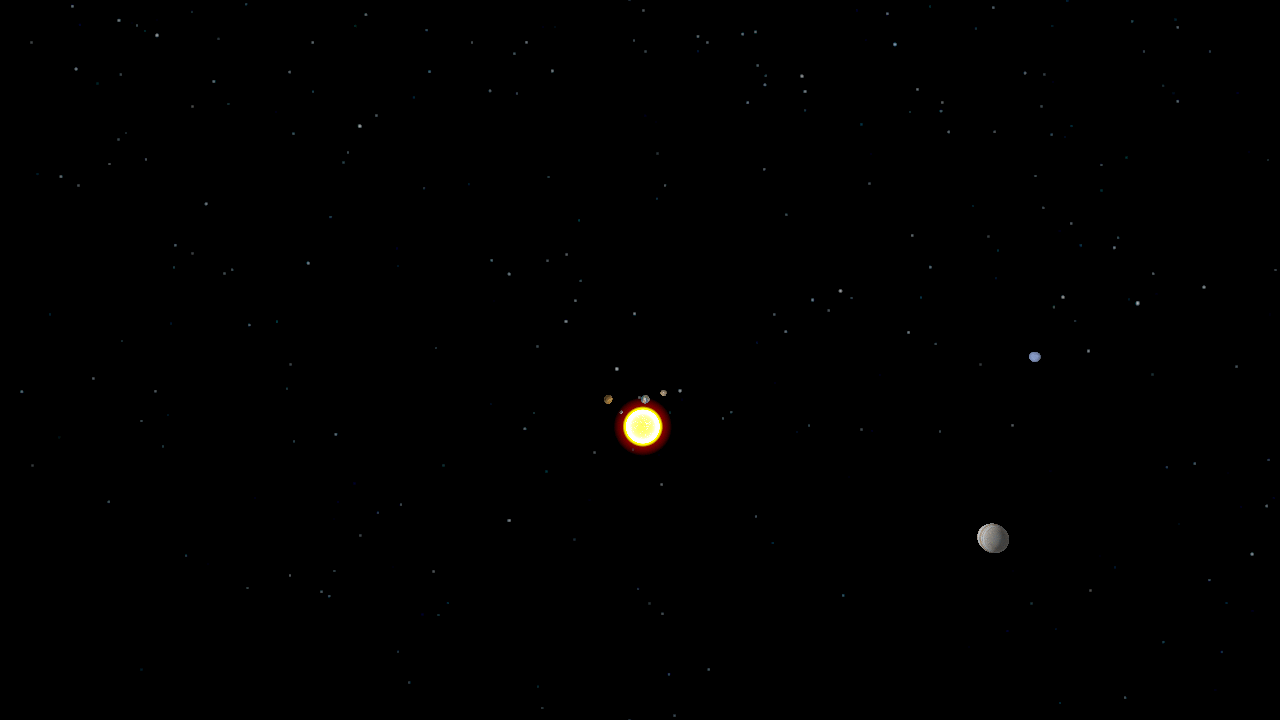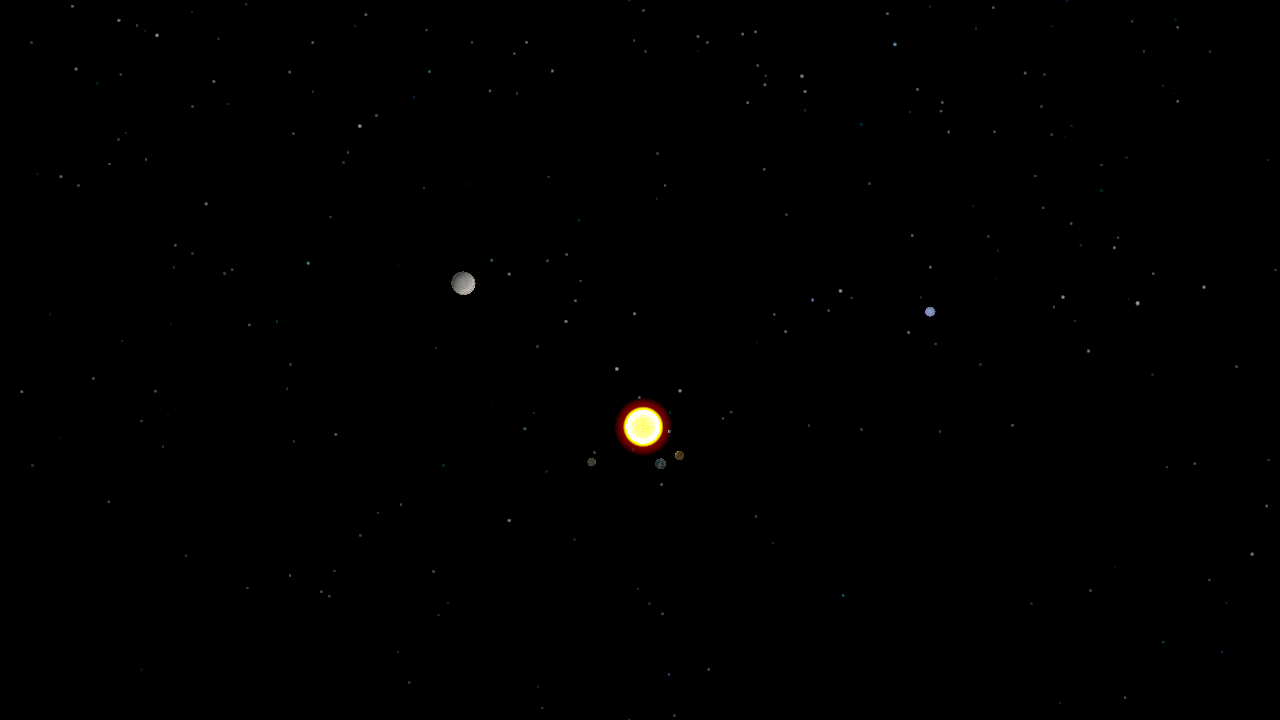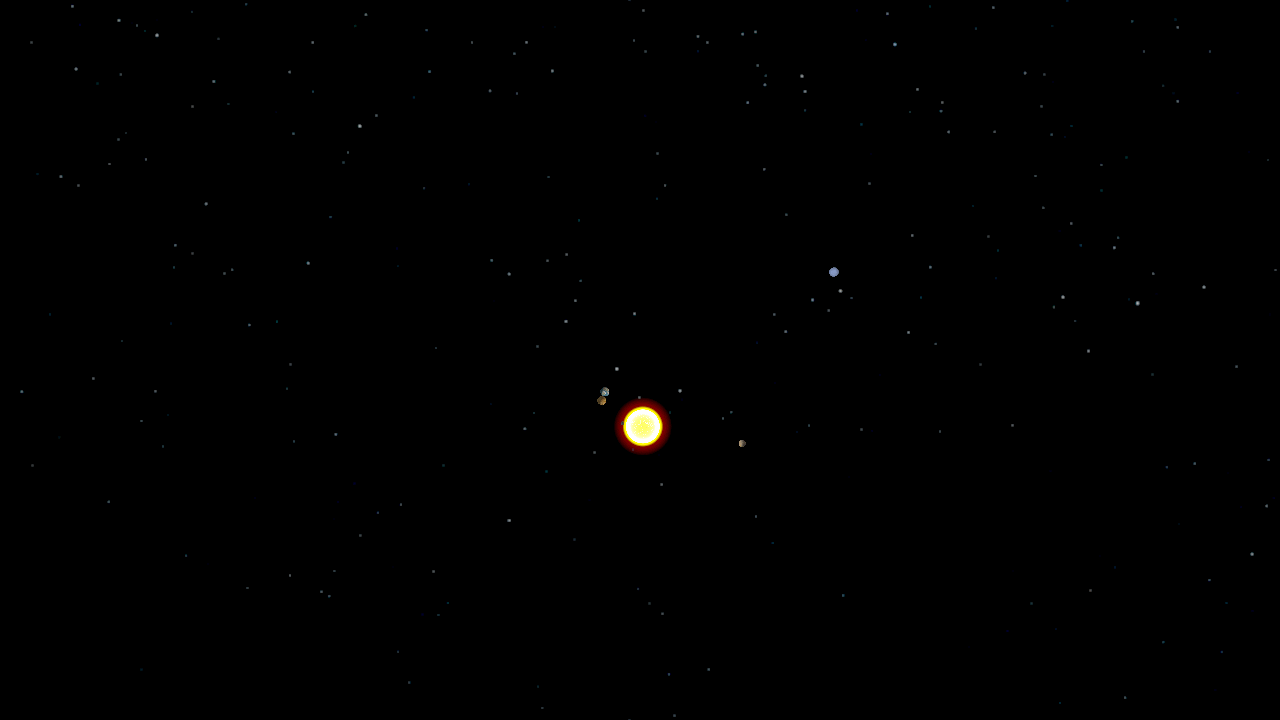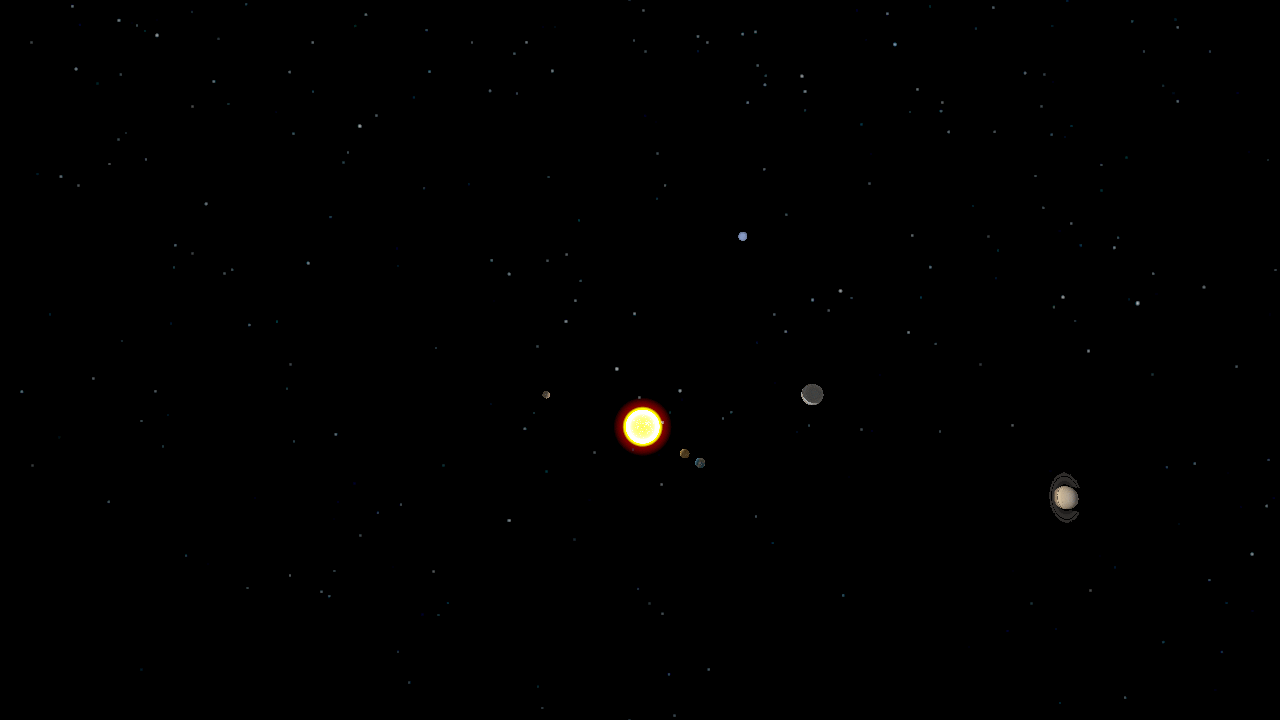 The undergraduate Programming Paradigms class at the University of Notre Dame
introduces undergraduate students to a variety of parallel and distributed
programming models. Work Queue is used as an example of large scale distributed computing.
Using a solar system simulator developed in a previous assignment,
students were tasked with splitting a trajectory
of the planets' positions into individual frames, populating POVRay
scene files, rendering the scenes in a distributed manner using Work
Queue, and combining the frames into a movie using ImageMagick. Since
the students had used Python extensively, they found it very easy to
write a single Work Queue master using the Python bindings.
Several of the students went above and
beyond the requirements by adding textures to the planets and
animating the movement of the camera. The students really enjoyed the
assignment while learning about the advantages and pitfalls
of distributed computing.
The undergraduate Programming Paradigms class at the University of Notre Dame
introduces undergraduate students to a variety of parallel and distributed
programming models. Work Queue is used as an example of large scale distributed computing.
Using a solar system simulator developed in a previous assignment,
students were tasked with splitting a trajectory
of the planets' positions into individual frames, populating POVRay
scene files, rendering the scenes in a distributed manner using Work
Queue, and combining the frames into a movie using ImageMagick. Since
the students had used Python extensively, they found it very easy to
write a single Work Queue master using the Python bindings.
Several of the students went above and
beyond the requirements by adding textures to the planets and
animating the movement of the camera. The students really enjoyed the
assignment while learning about the advantages and pitfalls
of distributed computing.- Ronald J. Nowling and Jesus A. Izaguirre, University of Notre Dame
Scaling Up Comparative Genomics with Makeflow
 The CoGe Comparative Genomics Portal
provides on-the-fly genomic analysis and comparative tools for nearly
20,000 genomes from 15,000 organisms and has become more and more
popular as genome sequence has become less expensive. The portal runs
about 10,000 workflows a month and needed a robust solution for
distributed computing of various workflows that range from simple to
complex. Using Makeflow,
the CoGe team is modularizing the workflows being run through CoGe, has
early wins in delivering value to the system by easily
monitoring/restarting workflows, and is now starting to work on
distributing computation across multiple types of compute resources.
The CoGe Comparative Genomics Portal
provides on-the-fly genomic analysis and comparative tools for nearly
20,000 genomes from 15,000 organisms and has become more and more
popular as genome sequence has become less expensive. The portal runs
about 10,000 workflows a month and needed a robust solution for
distributed computing of various workflows that range from simple to
complex. Using Makeflow,
the CoGe team is modularizing the workflows being run through CoGe, has
early wins in delivering value to the system by easily
monitoring/restarting workflows, and is now starting to work on
distributing computation across multiple types of compute resources.- Eric Lyons, University of Arizona
Applied Cyber Infrastructure Class at U. Arizona
 The Applied Cyber Infrastructure Concepts course at the University of Arizona makes use of the Cooperative Computing Tools
to teach
principles of large scale distributed computing. The project-based
course introduced fundamental concepts and tools for analyzing large
datasets on national computing resources and commercial cloud providers.
For the final project, students developed an appliance for annotating
multiple rice genomes using MAKER software, distributed across Amazon and FutureGrid resources via the Work Queue
framework. They also developed a web application to visualize the
performance and utilization of each cloud appliance in real-time,
overall progress of the rice annotation process, and integrated the
final results into a genome browser.
The Applied Cyber Infrastructure Concepts course at the University of Arizona makes use of the Cooperative Computing Tools
to teach
principles of large scale distributed computing. The project-based
course introduced fundamental concepts and tools for analyzing large
datasets on national computing resources and commercial cloud providers.
For the final project, students developed an appliance for annotating
multiple rice genomes using MAKER software, distributed across Amazon and FutureGrid resources via the Work Queue
framework. They also developed a web application to visualize the
performance and utilization of each cloud appliance in real-time,
overall progress of the rice annotation process, and integrated the
final results into a genome browser.- Nirav Merchant and Eric Lyons, University of Arizona
Global Access to High Energy Physics Software with Parrot and CVMFS
Scientists searching for the Higgs boson have profited from Parrot's new support for the CernVM Filesystem (CVMFS), a network filesystem tailored to providing world-wide access to software installations. By using Parrot, CVMFS, and additional components integrated by the Any Data, Anytime, Anywhere project, physicists working in the Compact Muon Solenoid experiment have been able to create a uniform computing environment across the Open Science Grid. Instead of maintaining large software installations at each participating institution, Parrot is used to provide access to a single highly-available CVMFS installation of the software from which files are downloaded as needed and aggressively cached for efficiency. A pilot project at the University of Wisconsin has demonstrated the feasibility of this approach by exporting excess compute jobs to run in the Open Science Grid, opportunistically harnessing 370,000 CPU-hours across 15 sites with seamless access to 400 gigabytes of software in the Wisconsin CVMFS repository.- Dan Bradley, University of Wisconsin and the Open Science Grid
Rapid Processing of LIDAR Data in the Field with Makeflow
 Makeflow is used to manage the data processing workflow of the Airborne Lidar Processing System (ALPS) for the Experimental Advanced Airborne Research Lidar (EAARL)
at the USGS. Over the course of several hours of flight time, about a
gigabyte of raw LIDAR waveform data is collected. This data must be
geo-referenced in order to convert it into point clouds broken into 2km
tiles suitable for GIS use. When we collect this data in the field, it
is critical that the field crew can process the data rapidly and take a
look at it for obvious problems so they can be corrected before the next
flight.
Makeflow is used to manage the data processing workflow of the Airborne Lidar Processing System (ALPS) for the Experimental Advanced Airborne Research Lidar (EAARL)
at the USGS. Over the course of several hours of flight time, about a
gigabyte of raw LIDAR waveform data is collected. This data must be
geo-referenced in order to convert it into point clouds broken into 2km
tiles suitable for GIS use. When we collect this data in the field, it
is critical that the field crew can process the data rapidly and take a
look at it for obvious problems so they can be corrected before the next
flight.Using Makeflow, the data can be processed quickly on a portable 32-core cluster in the field in about 20 minutes. The data can be processed fast enough to do some cursory analysis and also re-process it a few times if needed to troubleshoot issues. Using Makeflow, it is easy to run the exactly same workflow in the field on the portable cluster or back in the office on a multi-core system.
- David Nagle, US Geological Survey








