Recent News in the CCL
CCL Team at GCASR 2025
Members of the CCL team traveled to Chicago, Illinois on May 8 to attend GCASR 2025 (12th Greater Chicago Area Systems Research Workshop).
CCL team members presented their work in both poster sessions.
Barry Sly-Delgado presented: Task Graph Restructuring via Function Based Annotation For Large-Scale Scientific Applications.
Colin Thomas presented: Enabling Tailored Optimizations for Scientific Workflows through I/O Tracing.
Md. Saiful Islam presented: Floability: Enabling Portable Scientific Workflows Across HPC Facilities.
Jin Zhou presented: Effectively Exploiting Node-Local Storage For Data-Intensive Scientific Workflows.
Reshaping High Energy Physics Applications Using TaskVine @ SC24
Barry Sly-Delgado presented our paper titled: "Reshaping High Energy Physics Applications for Near-Interactive Execution Using TaskVine" at the 2024 Supercomputing Conference in Atlanta, Georgia. This paper investigates the necessary steps to convert long-running high-throughput high energy physics applications to high concurrency. This included incorporating new functionality within TaskVine. The paper presents the speedup gained as changes were incorporated to the workflow execution stack for application DV3. We eventually achieve a speedup of 13X.
Configurations for each workflow execution stack as improvements were made.
Starting with stack 1. The first change incorporated was to the storage system where initial data sets are stored. This change showed little improvement, taking 3545s runtime to 3378s. This change is minimal as much of the data handling during application execution is related to intermediate results. Initially, with stack 1 and 2, intermediate data movement is handled via a centralized manager.
data movement during application execution between Work Queue and TaskVine. With TaskVine, the most data exchanged between any two nodes tops off around 4GB (the manger is node 0). With Work Queue the most data transferred is 40GB
Incorporated in stack 3 is a change of scheduler, TaskVine. Here, TaskVine allows for intermediate results to be stored on node-local storage and transferred between peer compute nodes. This relieves strain on the centralized manager and allows it to schedule tasks more effectively. This change drops the runtime to 730s.
CDF of task runtimes within the application per execution paradigm. With Function Calls, individual tasks execute faster.
Our final improvement changes the previous task execution paradigm within TaskVine. Initially "PythonTasks" serialized functions along with arguments and distributed them to compute nodes to execute individual tasks. Under this paradigm, the python interpreter would be invoked for each individual task. Our new task execution paradigm, "Function Calls" stands up a persistent Python process, "Library Task", that contains function definitions that can be invoked via individual function calls. Thus, invocations of the Python interpreter are reduced from per-task to per-compute-node. This change reduces runtime to 272s for a 13X speedup from our initial configuration.
Application execution comparison between stack configurations.
Shepherd Paper at WORKS/SC 2024
Grad student Saiful Islam presented our paper on Shepherd at the 19th Workshop on Workflows in Support of Large-Scale Science at Supercomputing 2024 in Atlanta, Georgia.Shepherd is a local workflow manager that enables a fleet of actions and services to functionally behave as a single task. This allows us to seamlessly deploy these local workflows into HPC clusters at large scale. For example, consider a workflow for large-scale distributed drone simulation. This workflow includes steps such as preprocessing, creating a configuration, running simulations, and post-processing. We aim to deploy the "run simulation" step in HPC clusters at large scale. However, each "run simulation" step includes a local workflow that needs to perform specific actions and start services like Gazebo, PX4, and Pose Sender. These services and actions depend on each other’s internal states. For example, when Gazebo reaches the "ready" state, the nth PX4 service is started. Without Shepherd, the workflow becomes as shown in the following figure:
With Shepherd, the entire "run simulation" step becomes a single task of running Shepherd with a specified configuration. Shepherd starts the required actions and services, manages dependencies, and ensures the graceful shutdown of services when they are no longer needed. This enables complex workflows of actions and services to be seamlessly integrated with a larger distributed workflow manager, as shown in the following figure:
The paper discusses the architecture and design principles of Shepherd, showcasing its application to large-scale drone simulations and integration testing.
Shepherd uses a YAML-based workflow description to define tasks, dependencies, and execution conditions. It monitors logs and file generation to infer internal states and manage lifecycles effectively. Additionally, it generates three visualizations post-execution for debugging and documentation. For instance, the figure below illustrates a timeline of a 100-drone simulation distributed across 25 nodes, with each Shepherd instance managing 4 drones. A zoomed-in view highlights how components execute at varying times across nodes, which becomes challenging without awareness of service readiness. Shepherd simplifies this with YAML-based configurations and internal state tracking.
For all the details, please check out our paper here:
- Md Saiful Islam, Douglas Thain, Shepherd: Seamless Integration of Service Workflows into Task-Based Workflows through Log Monitoring, 19th Workshop on Workflows in Support of Large-Scale Science at ACM Supercomputing, pages 1-8, November, 2024.
Tue, 07 Jan 2025 23:50:00 +0000
Data Pruning Mechanism in Daskvine
In our recent work, we introduced a file pruning technique into DaskVine to address challenges in managing intermediate files in DAG-based task graphs. This technique systematically identifies and removes intermediate stale files—those no longer needed by downstream tasks—directly from worker nodes. By freeing up storage in real time, file pruning not only enables workers to process more computational tasks with limited disk space but also makes applications feasible that were previously constrained by storage limitations.
Specifically, the pruning algorithm monitors task execution in the graph, categorizing tasks as waiting or completed. Once a task finishes, it prunes its parent tasks’ output files if all dependent child tasks are done and immediately submits dependent tasks for execution when their inputs are ready.
To evaluate the effectiveness of our file pruning technique, we utilized four key metrics:
File Retention Rate (FRR): FRR represents the ratio of a file's retention time on storage to the total workflow execution time. It indicates how long files remain in storage relative to the workflow's progress. A shorter FRR reflects more efficient pruning. The following figures show the FRR over all intermediate files, each producing by one specific task. The left graph (FCFS) shows higher and more uniform FRR, where files remain on storage for a significant portion of the workflow. In contrast, the right graph (FCFS with Pruning) shows a clear reduction in FRR for most files, which proves that pruning reduces storage usage by removing stale files promptly.
Accumulated Storage Consumption (ASC) and Accumulated File Count (AFC): ASC measures the total amount of storage consumed across all worker nodes throughout the workflow execution, while AFC tracks the total number of files retained across all worker nodes during the workflow. In the following, the left graph shows that both ASC and AFC steadily increase throughout the workflow, peaking at 1082.94 GB and 299,787 files, respectively. This highlights significant storage pressure without pruning. The right graph demonstrates dramatic reductions, with peaks at 326.78 GB for ASC and 80,239 files for AFC. This confirms that pruning effectively reduces storage consumption and file count by promptly removing unnecessary intermediate files.
Worker Storage Consumption (WSC): WSC reflects the storage consumption of individual workers at any point during the workflow execution. It helps assess the balance of storage usage among workers. The left graph shows a peak WSC of 24.95 GB, indicating high storage pressure on individual workers. In contrast, the right graph shows a significantly lower peak of 7.07 GB, demonstrating that pruning effectively reduces storage usage per worker and balances the load across the cluster.
Mon, 09 Dec 2024 16:10:00 +0000
TaskVine + Parsl Integration
The Cooperative Computing Lab team has an ongoing collaboration with the Parsl Project, maintaining the TaskVine Executor for use with the Parsl workflow system. Using the TaskVine executor involves expressing an application using the Parsl API, where tasks are created and managed using the Parsl Data Flow Kernel. Tasks are passed to the executor which makes final scheduling decisions.
The TaskVine executor offers a number of features involving data management and locality scheduling. The distinction between the TaskVine executor and other available executors is largely the awareness of data dependencies and the use of node-local storage. There are a number of features to the TaskVine executor that have been recently added or are in progress.
One prominent new feature is the extension of TaskVine function invocations to the TaskVine executor. Function invocations reduce the overhead of starting new tasks, considerably benefiting workflows with many short-running tasks. TaskVine function calls have been described with greater detail in this article by Thanh Son Phung. This benefit to task startup latency is especially useful for the Parsl/TaskVine executor, since Parsl is a python-native application. Normally each task must consider starting a new python process and loading all modules used by Parsl as well as the user's application. With short running tasks, such as ones resembling quick function calls, the overhead of loading this information from shared storage, or transferring it to local storage and starting a new python process can be greater than the running time of the task itself. TaskVine function invocations allow this runtime environment information to be effectively cached, and multiple tasks to run sequentially in the same python process. A molecular dynamics application using the Parsl/TaskVine executor is shown below, with L1 being simple tasks, and L2 using TaskVine function invocations.
Other new changes to the TaskVine executor include the addition of the tune_parameters to the executor configuration. TaskVine is highly configurable by users to suit the needs of their particular application or cluster resources. Adding this option to the TaskVine executor allows users to automatically replicate intermediate data, configure the degree of replication desired, disable peer transfers, and several in-depth scheduling options which fit different specific task dispatch and retrieval patterns.
Accelerating Function-Centric Applications via Reusable Function Context in Workflow Systems
Modern applications are increasingly being written in high-level programming languages (e.g., Python) via popular parallel frameworks (e.g., Parsl, TaskVine, Ray) as they help users quickly translate an experiment or idea into working code that is easily executable and parallelizable on HPC clusters or supercomputers. Figure 1 shows the typical software stack of these frameworks, where users wrap computations into functions, which are sent to and managed by a parallel library as a DAG of tasks, and these tasks eventually are scheduled by an execution engine to execute on a remote worker node.A traditional way to execute functions remotely is to translate them into executable tasks by serializing functions and their associated arguments into input files, such that these functions and arguments are later reconstructed on remote nodes for execution. While this way fits function-centric applications naturally into well understood task-based workflow systems, it brings a hefty penalty to short-running functions. A function now takes extra time for its states to be sent and reconstructed on a remote node which are then unnecessarily destroyed at the end of that function’s execution.
At HPDC 2024, graduate students Thanh Son Phung and Colin Thomas proposed the idea that function contexts, or states, should be decoupled from function’s actual execution code. This removes the overhead of repeatedly sending and reconstructing a function’s state for execution, and allows functions of the same type to share the same context. The rest of the work then addresses how a workflow system can treat a function as a first-class citizen by discovering, distributing, and retaining such context from a function. Figure below shows the execution time of the Large-Scale Neural Network Inference application, totaling 1.6 million inferences separated into 100k tasks, with increasing levels of context sharing (L1 is no sharing, L3 is maximum sharing). Decoupling the inference function’s context from the inference massively reduces the execution time of the entire workflow by 94.5%, from around 2 hours to approximately 7 minutes.
An interested reader can find more details about this work in the paper:Thanh Son Phung, Colin Thomas, Logan Ward, Kyle Chard, and Douglas Thain. 2024. Accelerating Function-Centric Applications by Discovering, Distributing, and Retaining Reusable Context in Workflow Systems. In Proceedings of the 33rd International Symposium on High-Performance Parallel and Distributed Computing (HPDC '24). Association for Computing Machinery, New York, NY, USA, 122–134. https://doi.org/10.1145/3625549.3658663
Mon, 11 Nov 2024 16:07:00 +0000
A New Visualization Tool for TaskVine Released
We released a web-based tool to visualize runtime logs produced by TaskVine, available on Github. Using this tool involves two main steps. First, the required data in CSV format must be generated for the manager, workers, tasks, and input/output files. After saving the generated data in the directory, users can start a port on their workstation to view detailed information about the run. This approach offers two key advantages: the generated data can be reused multiple times, minimizing the overhead of regeneration, and users can also develop custom code to analyze the structured data and extract relevant insights.
For example, the first section describes the general information of this run, including the start/end time of the manager, how many tasks are submitted, how many of them succeeded or failed, etc.
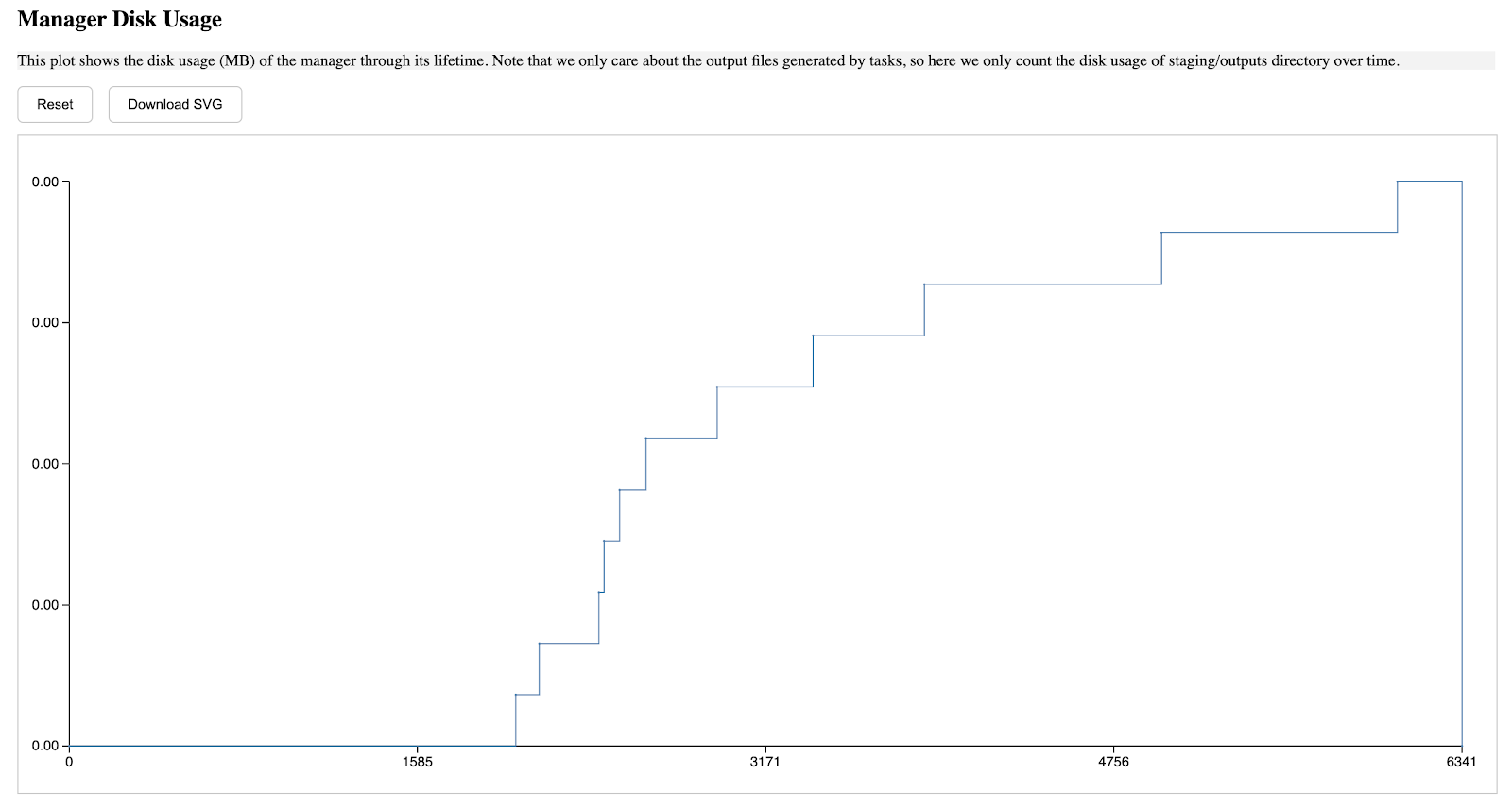
The second section describes the manager's storage usage through its lifetime, the a-axis starts from when the manager is started, and ends when the manager is terminated, the y-axis is in MB unit, and such pattern is applied to all diagrams in this report.
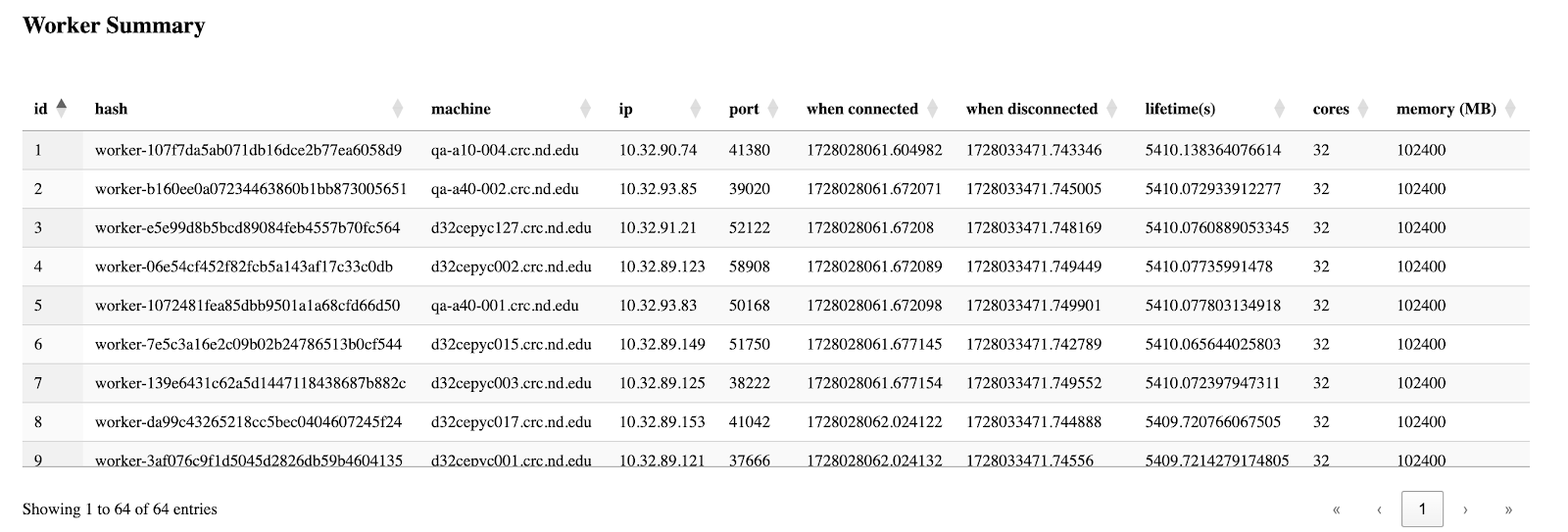
The third section is the table of all workers' information, which is basically grabbed from the csv files from the backend, but this enables users to easily sort by their interested columns.
The fourth section is the storage consumption among all workers. Several buttons in the top are provided, to turn the y-axis to a percentage unit, or to highlight one worker that is of interest.
The fifth section is the number of connected workers throughout the manager's lifetime, hovering the mouse on a point shows the information of a connected/disconnected worker.
The sixth section shows the number of concurrent workers throughout the manager's lifetime.
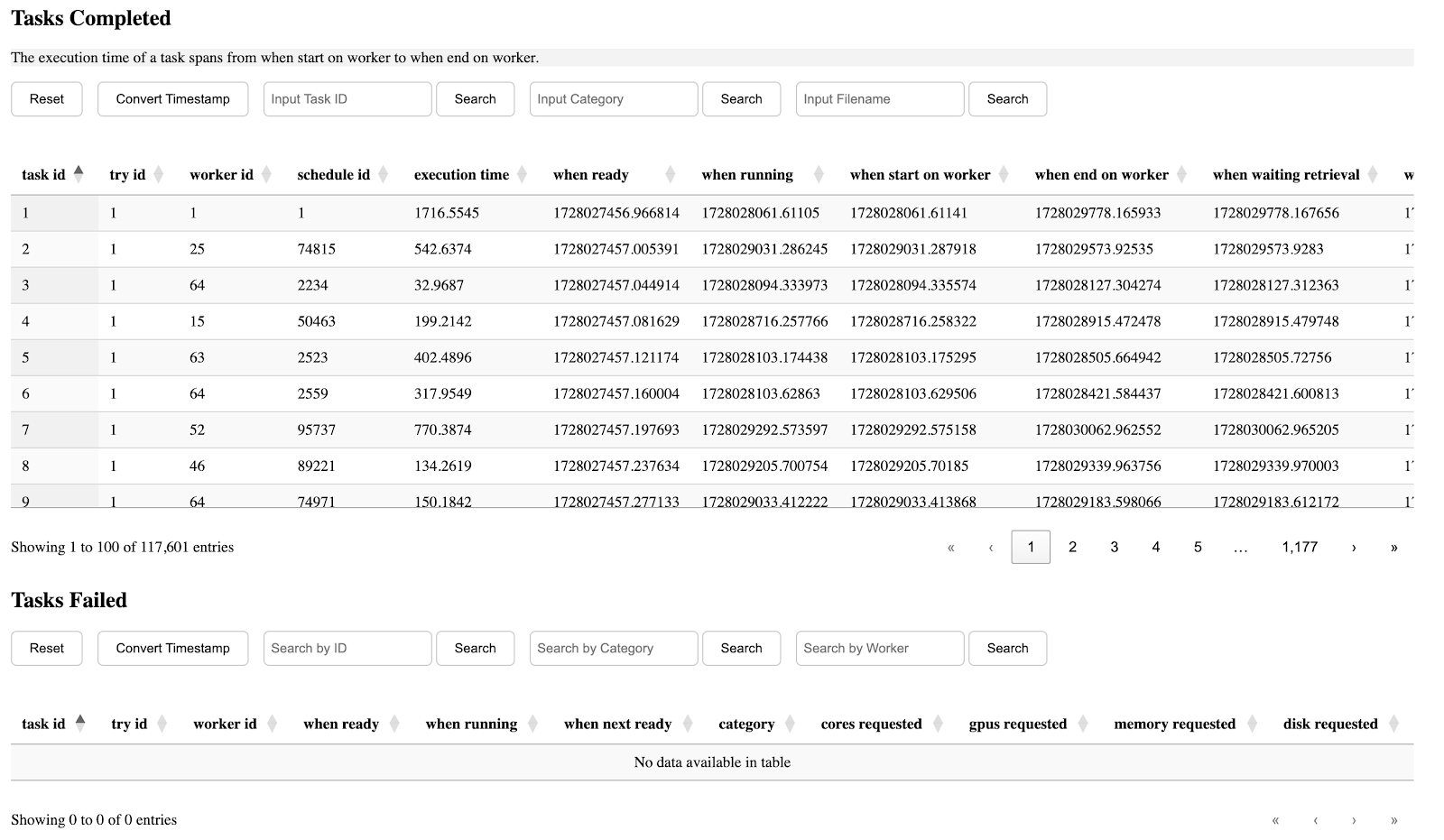
The seventh section is table of completed/failed tasks and their information.
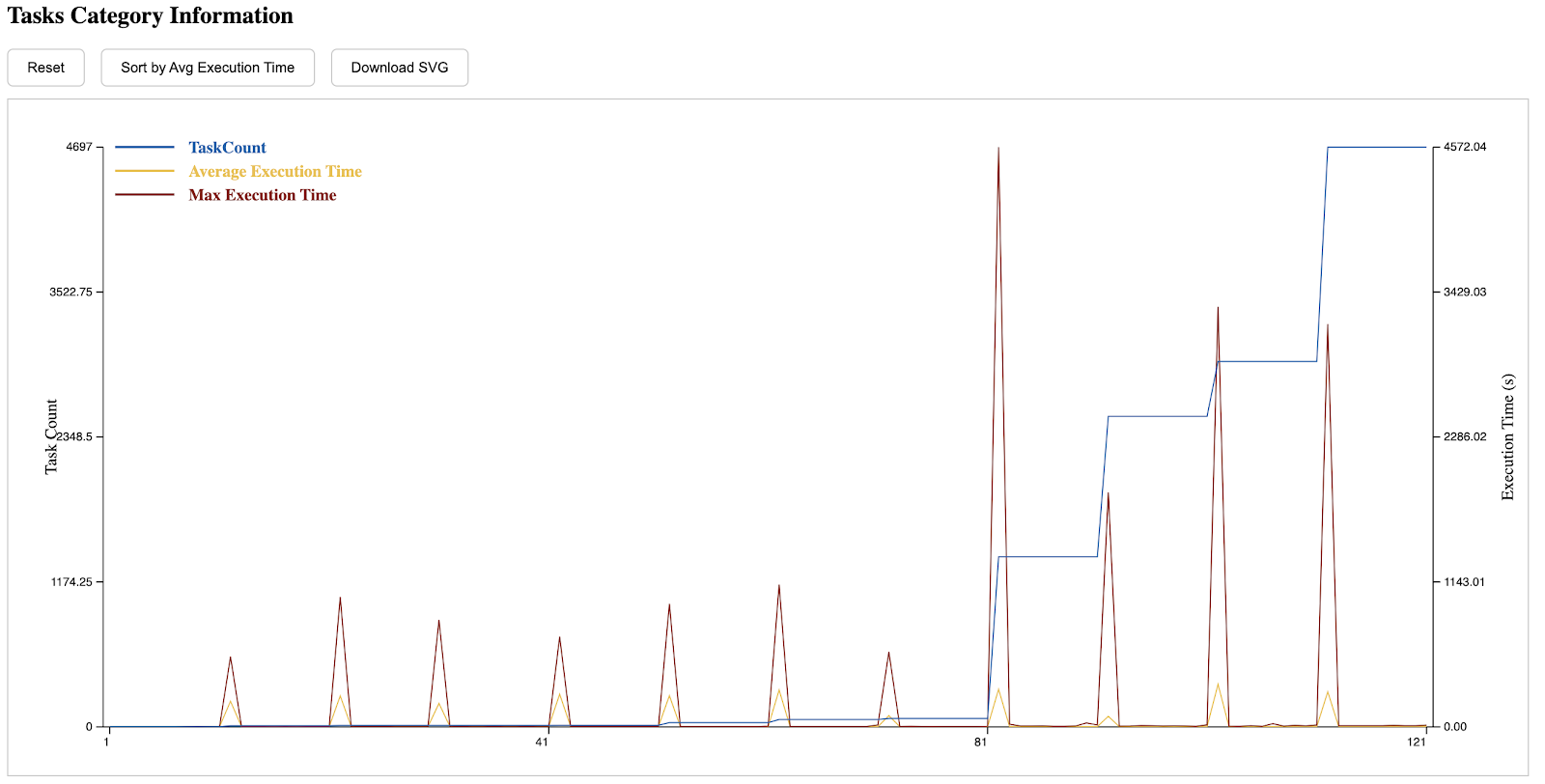
The eighth section is the execution time distribution of different tasks. Those with a lower index on the left side of the x-axis are submitted earlier, while those on the right are submitted later. A CDF can be seen by clicking the button on the top.
The tenth section demonstrates the general runtime distribution of tasks, the y-axis is the worker-slot pair. In the following example, we have 64 workers and each with 20 cores. One can zoom in the diagram and hover to see the detailed information of one task, which is particularly useful when examining outliers.
The last section is about the structure of the compute graphs. Nodes in the graph are tasks with an index label, while edges are the dependencies between input/output files of tasks. Weights on those task->file edges are the execution time of tasks, while those on the file->task edges are the waiting time starting from when a file is produced to when it is consumed by a consumer task.
This tool works well under the scale of hundreds of thousands of tasks, but for large runs, which may have millions of tasks, the online visualization tool may be unable to process such amount of data because the data transferring bottleneck between backend and frontend. Under such case, or just for convenience, we recommend the users to use tools under the pyplot directory, which is more lightweight and uses traditional matplotlib and seaborn to draw diagrams. Detailed explanations are provided under the README file.
Mon, 04 Nov 2024 23:34:00 +0000Integrating TaskVine with Merlin
Graduate student, Barry Sly-Delgado, completed a summer internship onsite at Lawrence Livermore National Laboratory where he worked on integrating TaskVine with Merlin, an executor for machine learning workflows. Barry worked as a member of the WEAVE team under Brian Gunnarson and Charles Doutriaux.
Previously, Merlin used Celery to distribute tasks across a compute cluster. With TaskVine's addition, utilization of in-cluster resources (bandwidth, disk) is available for workflow execution. Existing Merlin specifiacations can use TaskVine as a task scheduler with little change to the specification itself.
Merlin works with TaskVine by utilizing the Vine Stem, a DAG manager that borrows the concepts of groups and chains to create workflows from Celery. With this, the Vine Stem sends tasks (Merlin Steps) to the TaskVine manager for execution. Execution of these tasks eventually create a directory hierarchy that previous Merlin workflows already do. In addition to the workflow specification, a Merlin Specification contains specifications for starting workers, which are submitted via a batch system (HTCondor, UGE,Slurm)
Architecture of Merlin with TaskVine
Sample Merlin Specification Block With TaskVine as Task Server
The TaskVine task server option will be included in an upcoming release of Merlin. We would be happy to find more use cases so please check it out once released!
Wed, 16 Oct 2024 19:14:00 +0000
Introducing Shepherd: Simplifying Integration of Service Workflows into Task-Based Workflows
We are pleased to announce the release of Shepherd, an open-source tool designed to streamline the integration of service workflows into task-based workflows. Shepherd enables users to manage applications that require dynamic dependencies and state monitoring, ensuring that each component in a workflow starts only after its dependencies have reached specified states.
What Is Shepherd?
Shepherd is a local workflow manager and service orchestrator that launches and monitors applications according to a user-defined configuration. It bridges the gap between persistent services and traditional task-based workflows by treating services as first-class tasks. This approach allows for seamless integration of services that run indefinitely or until explicitly stopped, alongside actions that run to completion.
By analyzing log outputs and file contents, Shepherd tracks the internal states of tasks without the need to modify their original code. This capability is particularly useful when integrating persistent services into workflows where tasks may depend on the internal states of these services rather than their completion.
Key Features
- Services as Tasks: Treats both actions (tasks that run to completion) and services (persistent tasks that require explicit termination) as tasks within a workflow, allowing integration into task-based workflow managers.
- Dynamic Dependency Management: Initiates tasks based on the internal states of other tasks, enabling complex state-based dependencies. Supports both "any" and "all" dependency modes for flexible configurations.
- State Monitoring: Continuously monitors the internal state of each task by analyzing standard output and specified files, updating states in real-time based on user-defined patterns.
- Graceful Startup and Shutdown: Ensures tasks start only when their dependencies are met and provides controlled shutdown mechanisms to maintain system stability and prevent data loss.
- Logging and Visualization: Generates comprehensive logs and state transition data, aiding in performance analysis and debugging.
- Integration with Larger Workflows: By encapsulating service workflows into single tasks, Shepherd enables integration with larger distributed workflow managers, enhancing workflow flexibility and reliability.
Documentation and Source Code
For detailed documentation, examples, and source code, please visit the Shepherd GitHub Repository.
Mon, 14 Oct 2024 13:55:00 +0000
TaskVine at ParslFest 2024
On September 26-27 members of the CCL team attended ParslFest 2024 in Chicago, Illinois to speak about TaskVine and connect with our ongoing collaborators at the Parsl Project.
Grad student Colin Thomas delivered a talk titled "Parsl/TaskVine: Interactions between DAG Manager and Workflow Executor."
The talk highlighted recent developments in the implementation of TaskVine Temporary Files within the Parsl/TaskVine Executor. Temporary files allow users to express intermediate data in their workflows. Intermediate data are items produced by tasks, and consumed by tasks. With the use of TaskVine Temporary Files, this intermediate data will remain in the cluster for the duration of a workflow. Keeping this data in the cluster can benefit a workflow that produces intermediate files of considerable quantity or size.
In addition to adding temporary files to the Parsl/TaskVine executor, the talk presented ongoing work on the development of intermediate data-based "task-grouping" in TaskVine and extending it to the TaskVine executor. The concept of grouping related tasks may sound familiar to Pegasus users or other pre-compiled DAG workflow systems. Currently task-grouping is focused on scheduling sequences of tasks which contain intermediate dependencies. The desired outcome is for TaskVine to interpret pieces of the DAG to identify series of sequentially-dependent tasks in order to schedule them on the same worker, such that intermediate data does not need to travel to another host. Extending this capability to Parsl brought about an interesting challenge which questioned the relationship between DAG manager and executor.
The problem is that in the typical workflow executor scheme, such as with Parsl or Dask, the executor will receive tasks from the DAG manager only when they are ready to run. In order for TaskVine to identify sequentially dependent tasks it needs to receive tasks which are future dependencies of those that are ready to run.
The solution to this came about as a special implementation of a Data Staging Provider in Parsl, which is the mechanism for determining whether data dependencies are met. When the TaskVine staging provider encounters temporary files, it will lie to the Parsl DFK (Data Flow Kernel) in saying the file exists. Therefore Parsl will send all temporary-dependent tasks to TaskVine without waiting for the files to actually materialize. This offers TaskVine relevant pieces of the DAG to inspect and identify dependent sequences.
The impact of task grouping was evaluated on a benchmark application. Each benchmark run consists of 20 task sequences of variable length. Each task in a sequence produces and consumes an intermediate file of variable size. The effect on running time is shown while adjusting these parameters. Grouping tasks was shown to benefit the performance across a variety of sequence lengths, and when intermediate data size exceeds 500MB.
In addition to the positive benchmark results, this talk promoted discussion about this non-traditional interaction between DAG manager and executor. The DAG manager has insight into future tasks and their dependencies. The executor possesses knowledge about data locality. Combining this information to make better scheduling decisions proved to have some utility, yet it challenged the typical scheme of communication between Parsl and the TaskVine executor.
Predicting Resources of Tasks in Dynamic Workflows with Bucketing Algorithms at IPDPS 2024
Thanh Son Phung will present Adaptive Task-Oriented Resource Allocation for Large Dynamic Workflows on Opportunistic Resources at the International Parallel and Distributed Processing Symposium 2024.
Non-technical users running dynamic workflows usually have no knowledge of resource (e.g., cores, memory, disk, GPUs) consumption of tasks within a given workflow. However, this omission of information can bring substantial performance degradation as it is important for workflow execution engines to know the exact amount of resources each task takes and make scheduling decisions accordingly.
In the figure above, an application submits many invocations of two functions: f and g. The invocations are transformed into tasks through many layers and scheduled on worker nodes. Note that the resource specification of each task may be unknown.
Since a manual resource specification for each task is unreliable, an automated solution is required. Such solution needs to be (1) general-purpose (can work with any workflow), (2) prior-free (no prior information is used as workflows may change over each run), (3) online (collect information and run as the workflow runs), and (4) robust (performant on many distributions and unexpected changes). The below figure shows two workflows: ColmenaXTB (top row) and TopEFT (bottom row). Resource types range from core, memory, disk, and execution time from left to right.
To predict resources of each task, we first collect resource records of completed tasks, group them into buckets of tasks based on their similarity in resource consumption, and probabilistically choose a bucket to allocate the next task, as shown in the example below.
We introduce two bucketing algorithms, namely Greedy Bucketing and Exhaustive Bucketing, which share the same principle of resource prediction and only differ in the way buckets of tasks are computed.
We compare the two algorithms with five alternative algorithms on 7 workflows with 3 resource types. Results show that Greedy Bucketing and Exhaustive Bucketing consistently outperform other algorithms by yielding the highest resource efficiency in the majority of cases.
Further details can be found in the paper:
- Thanh Son Phung, Douglas Thain, Adaptive Task-Oriented Resource Allocation for Large Dynamic Workflows on Opportunistic Resources, IEEE International Parallel and Distributed Processing Symposium (IPDPS), pages 12, May, 2024.
TaskVine at the HEP Analysis Grand Challenge
Barry Sly-Delgado and Ben Tovar recently presented our work on transforming high energy physics data analysis applications into near-interactive execution at the IRIS-HEP Analysis Grand Challenge Demo Day.
By using the TaskVine framework, we have enabled the transformation of a production physics data analysis application (DV3), reducing execution time from over an hour to just over three minutes
on about four thousand cores of a campus HPC cluster.
Advancements to hardware and software components in an application stack aid in the facilitation of application reshaping. With application reshaping, we are transitioning applications from long running to near-interactive. While improvements to hardware and low-level software improvements can produce measurable speedup, significant speedup is obtainable via optimization to the scheduling and execution layers of the distributed application.
TaskVine, our latest workflow technology, makes use of in-cluster bandwidth and disk to mitigate data movement by enabling peer transfers between worker nodes within a compute cluster. TaskVine is currently used to run a variety of custom data analysis workflows written by the CMS physics group at Notre Dame. These applications are written in high-level Python, making use of standard technologies like Numpy, Coffea, and Dask in order to generate large task graphs. TaskVine then takes that graph and deploys it into the cluster.
Our previous versions of these physics applications utilized our earlier WorkQueue executor. TaskVine improves upon the earlier system in two distinct ways:
First - While Work Queue only transfers data between the manager and workers, TaskVine transfers data directly between peer workers. Peer transfers relieve pressure on the manager to distribute data dependencies to worker nodes within the cluster. For workflows that generate large amounts of intermediate data, this can be extremely costly to scheduling performance, because the manager sends and receives large amounts of data.
This animation shows the data transfer throughout an application when using Work Queue:
Second - TaskVine extends the notion of task execution into a serverless model. Python code in the analysis application can be defined once as a LibraryTask which is deployed into the cluster. Then, lightweight FunctionCall tasks can invoke the code in the LibraryTask without constantly restarting the Python environment. In this modem the python interpreter is now invoked once per worker.
This graph compares the distribution of execution times between normal tasks and serverless function calls, improving the lower limit of execution time by an order of magnitude.
These four graphs show the overall performance of the data analysis application through four improvement states: first with Work Queue and Hadoop, then with Work Queue and VAST (new HPC filesystem), then TaskVine with peer transfers, and finally TaskVine with serverless execution. Overall, this transforms a one hour workflow into a 3 minute workflow.
Mon, 04 Mar 2024 14:39:00 +0000
CCTools 7.8.0 released
We are pleased to announce the release of version 7.8.0 of the
Cooperative Computing Tools from the University of Notre Dame, including
TaskVine, Work Queue, Makeflow, Parrot, Chirp, and other software.
Docs: https://cctools.readthedocs.io/
Download: http://ccl.cse.nd.edu/software/download
Forum: http://ccl.cse.nd.edu/community/forum
This is a feature release that primarily improves the scalability and performance of TaskVine:
Main Features:
TaskVine dask serverless execution (i.e., function calls instead of tasks) to reduce overheads, by @BarrySlyDelgado
TaskVine improvements to library and remove function calls dispatch, by @tphung3
TaskVine temporary file replication, by @colinthomas-z80
TaskVine automatic deletion of ancillary files (unlink_when_done) during workflow execution, by @btovar
TaskVine use of memory buffers to serialize functions, by @btovar
TaskVine cleanup of factory, worker information and stats, by @dthain
Fix TaskVine memory leak in python bindings, by @JinZhou5042
Improved Parsl integration with TaskVine and documentation, by @colinthomas-z80
Full Changelog
Distant Futures at SC 2023
Grad student Barry Sly-Delgado presented his recent work on "Minimizing Data Movement Using Distant Futures" at the research poster session at Supercomputing 2023:
Futures are a widely used concept for organizing concurrent computations. A Future is a variable that represents the result of a function call whose computation may still be pending. When evaluated, the future blocks (if needed) until the result becomes available. This allows for the easy construction of highly parallel programs.
Barry developed a future based execution model for the TaskVine workflow execution system. This allows one to submit and chain function calls written in plain Python. TaskVine then schedules the tasks throughout the cluster, providing a variety of data management services.
However, at very large scales, a natural bottleneck is the return of computed values back to the original manager. To overcome this, we introduce the notion of Distant Futures in which a value is not only pending in time, but potentially left in the cluster on a remote node. Tasks requiring this value can then be scheduled to the same node, or to transfer the value within the cluster, rather than bringing it back home.
Combining distant futures with asynchronous transfer provides significant benefits for applications that are bottlenecked in data transfer, as our results show:
For more details, check out the poster and short paper here:
- Barry Sly-Delgado and Douglas Thain, Poster: Minimizing Data Movement Using Distant Futures ACM/IEEE Supercomputing, November, 2023.
Maximizing Data Utility at HPPSS/SC 2023
Thanh Son Phung presented Maximizing Data Utility for HPC Python Workflow Execution at the High Performance Python for Science at Scale workshop at Supercomputing 2023.
This paper describes how the Parsl workflow system integrates with the TaskVine workflow executor in order to run complex Python environments at scale. Parsl provides a function-oriented programming model, generates a large number of tasks to be executed in a graph, and then passes the tasks down to TaskVine for execution on a cluster:
A significant challenging in scaling up Python applications is managing the large body of dependencies in the form of Python modules, system libraries, and so forth, that must be loaded every time a process starts. At very large scales, the cost of pulling all of these resources from a shared parallel filesystem can become a significant bottleneck. (Typically, parallel filesystems are optimized for the delivery of a few large files, rather than a large number of small files.)
TaskVine addresses the problem by exploiting the storage capability of each execution node, which is typically equipped with a local SSD for temporary storage. The TaskVine workers pull data into local storage, and then share files with each other (at the direction of the manager), resulting in less pressure on the shared filesystem, and overall speedups that improve with scale.
This technique was evaluated on a machine learning application requiring a (quite common) 4.4GB of software scaling up from 32 nodes x 16 cores to 256 nodes x 16 cores on a campus HPC cluster. At small scales, TaskVine provides a modest improvement, but as scale increases, the separation becomes more distinct:
The distribution of task execution times shows that the TaskVine variants (bottom row) shows that tasks run more consistently with fast individual execution times, once data is distributed across the cluster.
For the full details, see our paper here:
|
TaskVine Paper at WORKS/SC 2023
Barry Sly-Delgado presented our overview paper on TaskVine at the Workshop on Workflows in Support of Large Scale Science at Supercomputing 2023 in Denver, Colorado.
TaskVine is a system for executing data intensive workflows on large clusters. These workflows ma consist of thousands to millions of individual tasks that are connected together in a graph structure like this:
When executing this sort of workflow in a cluster, the movement of data between tasks is often the primary bottleneck, especially if each item must flow back and forth between a shared filesystem.
The key idea of TaskVine is to exploit the local storage available on each node of a cluster to relieve much of the load placed on a shared filesystem. A running TaskVine system consists of a manager process that coordinates data transfers between workers, like this:
A workflow in TaskVine is expressed in in Python by declaring the data assets needed by the workflow as file objects, and then connecting them to the namespace of each task to execute:
When the tasks execute on each worker, the various data objects are cached on local storage, and then linked into the execution sandbox of each running task. This provides the opportunity for data to be shared between tasks without moving data to/from the shared filesystem:
This paper gives three examples of applications constructed using TaskVine. TopEFT is a physics data analysis application built using Coffea+TaskVine, Colmena-XTB is a molecular modeling application built using Parsl+TaskVine, and BGD is a model training application built using TaskVine alone. Each of these codes is able to scale to 400-1000 nodes running on 1000-27000 cores total.
For all the details, please check out our paper here:
- Barry Sly-Delgado, Thanh Son Phung, Colin Thomas, David Simonetti, Andrew Hennessee, Ben Tovar, Douglas Thain, TaskVine: Managing In-Cluster Storage for High-Throughput Data Intensive Workflows, 18th Workshop on Workflows in Support of Large-Scale Science, November, 2023. DOI: 10.1145/3624062.3624277
CCTools 7.7.0 Released
We are pleased to announce the release of version 7.7. of the Cooperative Computing Tools from the University of Notre Dame, including TaskVine, Work Queue, Makeflow, Parrot, Chirp, and other software.
- Docs: https://cctools.readthedocs.io/
- Download: http://ccl.cse.nd.edu/software/download
- Forum: http://ccl.cse.nd.edu/community/forum
This is a feature release that primarily improves the scalability and performance of TaskVine:
Main Features:
- TaskVine workers now perform transfers asynchronously while communicating with manager, by @BarrySlyDelgado
- TaskVine Serverless function execution simplified and accelerated by @tphung3
- Improved integration of Parsl and TaskVine by @tphung3
- Improved scheduler performance in Work Queue and TaskVine by @colinthomas-z80
- Multiple improvements to reliability at scale with physics applications by @btovar
- Change to API for cancelling tasks: all tasks are now returned by
vine_waitby @dthain - Change to serverless resource management: functions consume library resources by @dthain
- Improved visualization by @JinZhou5042
- New environment.yml for development dependencies by @thieber22
Full Changelog: nightly...release/7.7.0
Tue, 03 Oct 2023 17:15:00 +0000CCTools 7.6.1 Released
We are pleased to announce the release of version 7.6.1
of the Cooperative Computing Tools from the University of Notre Dame,
including TaskVine, Work Queue, Makeflow, Parrot, Chirp, and other software.
Docs: https://cctools.readthedocs.io/
Mon, 24 Jul 2023 20:06:00 +0000
Download: http://ccl.cse.nd.edu/software/download
Forum: http://ccl.cse.nd.edu/community/forum
This release introduces several bug fixes and minor improvements to the TaskVine workflow executor and Work Queue scheduler.
TaskVine:
Forum: http://ccl.cse.nd.edu/community/forum
This release introduces several bug fixes and minor improvements to the TaskVine workflow executor and Work Queue scheduler.
TaskVine:
- Fix bug in task scheduling that would result in manager hanging. (Colin Thomas)
- Fix bug in execution of Dask task graphs that would lead to a hang. (Ben Tovar)
- Fix bug in deployment of serverless LibraryTask. (Thanh Phung)
- Add option to specify Python package dependencies inline. (Barry Sly-Delgado)
- Add visualization of task graph and documentation. (Douglas Thain)
Work Queue:
- Fix bug in execution of Dask task graphs that would lead to a hang. (Ben Tovar)
- Fix bug in deployment of serverless LibraryTask. (Thanh Phung)
- Add option to specify Python package dependencies inline. (Barry Sly-Delgado)
- Add visualization of task graph and documentation. (Douglas Thain)
Work Queue:
- Fix bug in task scheduling that would result in manager hanging. (Colin Thomas)
CCTools 7.6.0 Released
We are pleased to announce the release of version 7.6.0
of the Cooperative Computing Tools from the University of Notre Dame,
including TaskVine, Work Queue, Makeflow, Parrot, Chirp, and other software.
- Docs: https://cctools.readthedocs.io
- Download: http://ccl.cse.nd.edu/software/download
- Forum: http://ccl.cse.nd.edu/community/forum
This release introduces a number of performance, usability, and
documentation improvements to the TaskVine workflow executor.
TaskVine -
- Integration with Parsl workflow system. (Thanh Phung)
- Integration with Dask workflow system (Ben Tovar)
- Optimized scheduling performance with large numbers of workers. (Colin Thomas)
- Enhanced dispatch performance for small tasks. (Colin Thomas)
- vine_submit_workers combined tool for submtting workers. (Thanh Phung)
- Improved generation of file cache names (Barry Sly-Delgado)
- fetch_file interface to obtain cached files in the cluster. (Douglas Thain)
- Worker-to-worker authentication for peer transfers.
- Updated documentation. (All)
Work Queue:
- Enhanced dispatch performance for small tasks. (Colin Thomas)
Makeflow:
- Now supports TaskVine as an execution mode. (Douglas Thain)
General:
- TCP catalog updates are performed in a background process to reduce latency.
Thanks goes to the contributors for many features, bug fixes, and tests:
Thanh Son Phung
Barry Sly-Delgado
Colin Thomas
Ben Tovar
Douglas Thain
Enjoy!
Intro to TaskVine at GCASR 2023
Prof. Thain gave the afternoon keynote (Data Intensive Computing with TaskVine) at the GCASR Workshop in Chicago on April 24th. TaskVine is our latest workflow execution system, just released in March 2023.
The key idea of TaskVine is to exploit the local storage already embedded into clusters for use by complex workflows:
To accomplish this, each node runs a general purpose worker process that manages cached data under unique names, and runs each task within a private namespace:
You can write workflows directly to the TaskVine API, declaring files and tasks and connecting them together. Or, TaskVine can be used as an executor under existing workflow systems:
CCTools version 7.5.2 released
The
Cooperative Computing Lab is pleased to announce the release of version
7.5.2 of the Cooperative Computing Tools including TaskVine, WorkQueue,
Makeflow, Poncho, Chirp, JX, Parrot, and other software.
The software may be downloaded here:
http://ccl.cse.nd.edu/software/download
This is our first release of TaskVine, our new data intensive workflow execution system. TaskVine make extensive use of in-cluster storage to accelerate workflows, so that data "grows like a vine" through the cluster. See our extensive documentation and examples to learn more!
Thanks goes to the contributors for many features, bug fixes, and tests:
Mon, 17 Apr 2023 16:35:00 +0000
The software may be downloaded here:
http://ccl.cse.nd.edu/software/download
This is our first release of TaskVine, our new data intensive workflow execution system. TaskVine make extensive use of in-cluster storage to accelerate workflows, so that data "grows like a vine" through the cluster. See our extensive documentation and examples to learn more!
Detailed items in this release:
- [TaskVine] First official release (Everyone!)
- [General] Change catalog update to tcp by default (D. Thain)
- [Work Queue] Specify manager as an argument when using the factory in python (Ben Tovar)
- [Work Queue] New dynamic resource allocation strategies (Thanh Phung)
Thanks goes to the contributors for many features, bug fixes, and tests:
- Matthew Carbonaro
- Jachob Dolak
- Joseph Duggan
- Kelci Mohrman
- Thanh Son Phung
- David Simonetti
- Barry Sly-Delgado
- Douglas Thain
- Colin Thomas
- Ben Tovar
TaskVine System Architecture
TaskVine is our newest framework for building large scale data intensive dynamic workflows. This is the second in a series of posts giving a brief introduction to the system.
A TaskVine application consists of a manager and multiple workers running in a cluster. The manager is a Python program (or C if you prefer) that defines the files and tasks making up a workflow, and gives them to the TaskVine library to run. Both the files and the tasks are distributed to the workers and put together in order to produce the results. As a general rule, data is left in place on the workers wherever possible, so that it can be consumed by later tasks, rather than bringing it back to the manager.The workflow begins by declaring the input files needed by the workflow. Here, "file" is used generally and means any kind of data asset: a single file on the filesystem, a large directory tree, a tarball downloaded from an external url, or even just a single string passed from the manager. These files are pulled into the worker nodes on the cluster as needed. Then, the manager defines tasks that consume those files. Each task consumes one or more files, and produces one or more files.
A file declaration looks like one of these:
a = FileURL("http://server.edu/path/to/data.tar.gz")
b = FileLocal("/path/to/simulate.exe")
c = FileTemp()
And then a task declaration looks like this:
t = Task("simulate.exe -i input.dat")
t.add_input(a,"simulate.exe")
t.add_input(b,"input.dat")
t.add_output(c,"output.txt")
Tasks produce files which get consumed by more tasks until you have a large graph of the entire computation that might look like this:
To keep things clean, each task runs in a sandbox, where files are mapped in as names convenient to that task. As tasks produce outputs, they generally stay on the worker where they were created, until needed elsewhere. Later tasks that consume those outputs simply pull them from other workers as needed. (Or, in the ideal case, run on the same node where the data already is.).
A workflow might run step by step like this:
You can see that popular files will be naturally replicated through the cluster, which is why we say the workflow "grows like a vine" as it runs. When final outputs from the workflow are needed, the manager pulls them back to the head node or sends them to another storage target, as directed.
A Preview of TaskVine
We have seemed a bit quiet in the Cooperative Computing Lab lately, as we have been focused on building TaskVine, our new system for executing dynamic data intensive workflows.
I am excited that TaskVine is up and running in the lab now! We are working on refining a few technical issues and completing documentation and examples before making a general release. I would like to share some of the details and philosophy of the system as a preview before general availability.
TaskVine builds upon our many years of experience building large scale workflow systems that manage millions of jobs running on thousands of nodes in clusters, clouds, and grids. Some of our prior systems include Makeflow, which enabled the construction of large (static) task graphs to run on batch systems; Work Queue, which enabled the creation of applications that define and consume tasks dynamically; and Prune, which enabled the management of reproducible and shareable computations. (And you can still use those systems if you like.)
TaskVine has some familiar concepts: it too allows the creation of large dynamically parallel distributed applications. But where it really shines is in data management.
In TaskVine, data is first class citizen alongside tasks. Large data sets, software packages, and software services are explicitly declared and put into a working cluster where they can be replicated and shared among nodes. As an application runs, source data and derived results stay in the cluster and gradually creep from node to node, accelerating future tasks. Even later workflows can take advantage of data prepared or produced by prior workflows. We say that the workflow "grows like a vine" through the cluster.
The upshot is that TaskVine takes load off of the facility shared filesystem, which is often the bottleneck in scaling up large applications. Big software stacks, reference datasets, and intermediate steps are made accessible on the worker nodes in a disciplined way, rather than a thousand nodes pounding the same filesystem at once.
Of course, to make all this happen, there are a lot of fine details. In the next few posts, I'll give you an overview of the system architecture, and the new capabilities that we are building out.
Landlord Container Paper in TPDS 2023
Our latest work on container management was recently accepted to IEEE TPDS:
LANDLORD: Coordinating Dynamic Software Environments to Reduce Container Sprawl
his paper is the result of a continuing collaboration between the CCL at Notre Dame and the Parsl group at University of Chicago, led by Dr. Kyle Chard. Recent PhD grad Tim Shaffer led this work as part of a DOE Computational Science Graduate Fellowship, and current PhD student Thanh Phung joined the project and helped us to view the problem from a clustering perspective.
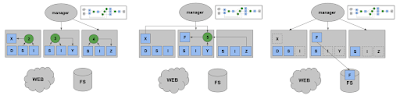
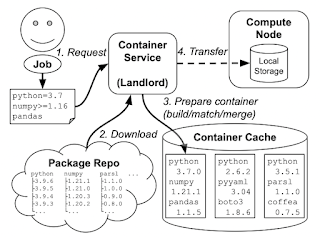
The problem is this: a variety of online services (like Binder, FuncX, and others) generate container images from software environment specifications, like a list of Conda packages. These images are used to execute code on clusters: sometimes long-running code like simulations, but also sometimes short bits of code, maybe even a single Python function call. If every user of the system asks for a slightly different software environment, then the system will quickly run out of space from those large container environments. ("container sprawl") So, we need some entity to manage the space and decide what containers to generate from packages, and which ones to delete:
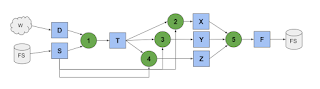
We observe that multiple requests might be satisfied by the same container. For example, these three jobs all express some constraints on packages A and B. (And one of those jobs doesn't care about the version at all. If we figure out the overlap between those requests, we can just use a single image to satisfy all three:
Going further, we can view this as an incremental online clustering problem. At any given moment, there are a certain number of containers instantiated. If a request matches one already, that's a hit and we just use it. Otherwise that's a miss and we have two choices: either insert a brand new container image that matches the request exactly, or merge an existing container with some new packages in order to satisfy the request. We decide what to do based on a single parameter alpha, which is a measure of distance between the request and the existing container.
Ok, so now how do we pick alpha? We evaluated this by running the Landlord algorithm through traces of software requests from high energy physics workloads and a public trace of binder requests.
There are two extremes to avoid: if alpha is very small, then we end up with a small number of very large containers. This is good for managing the shared cache, but bad because we have to keep moving very large containers out to the compute nodes. (And they may not fit!). On the other hand, if alpha is very large, then we end up with a large number of small containers. This results in a lot of duplication of packages, so the shared cache fills up faster, but the images are small and easy to move around.
As in most computer systems, there isn't one single number that is the answer: rather, it is a tradeoff between desiderata. Here, we have a broad sweet spot around alpha=0.8. But the good news is that the system has a broad "operational zone" in which the tradeoff is productive.
Sound interesting? Check out the full paper:
- Tim Shaffer, Thanh Son Phung, Kyle Chard, and Douglas Thain, LANDLORD: Coordinating Dynamic Software Environments to Reduce Container Sprawl, IEEE Transactions on Parallel and Distributed Systems, to appear in 2023. DOI: 10.1109/TPDS.2023.3241598
Mufasa: Robust Meta-Workflow Management at eScience 2022
This week, I'll be presenting Ben Lyon's M.S. project work on Mufasa at the eScience 2022 conference in Salt Lake City. Mufasa is a meta-workflow manager that schedules multiple workflows onto a cluster. When executing a large number of workflows, the WMS itself can have substantial resource consumption on the head node, network, and shared filesystem as it moves data to and from the cluster. Mufasa schedules these resources, ensuring that the system maintains throughput even under heavy load of arriving workflows.
- Ben Lyons and Douglas Thain, Robust Meta-Workflow Management with Mufasa, IEEE International Conference on eScience, October, 2022. [Short Paper] [Poster Image]